Dissecting the Circadian Clock in Real Time
Pushed into a new ‘time zone,’ aquatic organism’s clock components reveal surprising roles
As our bodies and minds continue to adjust to the recent time change, debates continue around society about whether to make daylight saving time a permanent fixture, eliminate it or stay with the current semi-annual clock adjustment.
As those discussions continue, scientists at the University of California San Diego and their colleagues have made progress in understanding the circadian clock, the 24-hour cycle that synchronizes with light-dark exposure, and how it functions (scientists in circadian and sleep research recommend permanent standard time as the healthiest option when considering light and dark exposure).
Internal biological clocks exist throughout the tree of life, rhythmically influencing daily activities and behavior. Two years ago a multi-institutional team of researchers assembled a circadian clock in a test tube for the first time to probe the components of the clock’s rhythms and interactions.
The “In Vitro Clock” helped the researchers analyze how the components of the clock interact in different times of the daily circadian cycle to control gene expression.
A new study led by UC San Diego and UC Merced researchers has expanded on this foundation with the development of a method to study how the circadian clock synchronizes with the environment in real time. As described in the journal Proceedings of the National Academy of Sciences, real-time capability allowed them to explore deeper into the clock’s previously unknown internal functions, including how time-setting signals are transmitted from its core—known as the oscillator—to the expression of genes that ensure a properly functioning clock.
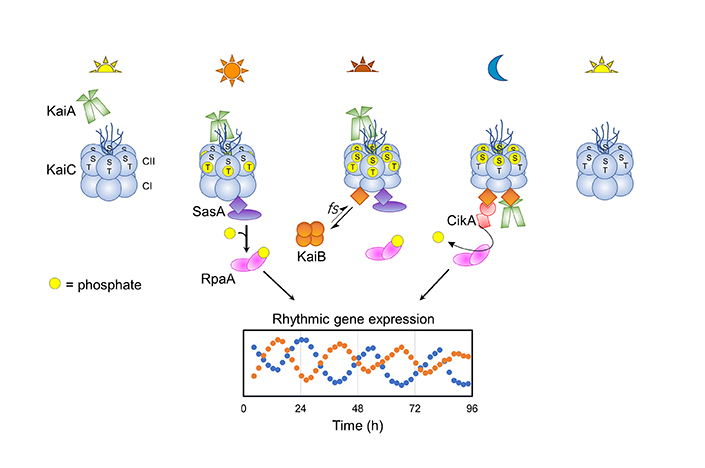 The circadian clock controls the rhythmic expression of hundreds of genes in the cyanobacterium Synechococcus elongatus. The pacemaker of the clock is a core oscillator made up of three proteins: KaiA, KaiB and KaiC. Temporal information is relayed to a transcription factor, RpaA, via two kinases, CikA and SasA, that physically interact with the Kai complex.
The circadian clock controls the rhythmic expression of hundreds of genes in the cyanobacterium Synechococcus elongatus. The pacemaker of the clock is a core oscillator made up of three proteins: KaiA, KaiB and KaiC. Temporal information is relayed to a transcription factor, RpaA, via two kinases, CikA and SasA, that physically interact with the Kai complex.
Postdoctoral Scholar Mingxu Fang and Professor Susan Golden in the School of Biological Sciences, along with their colleagues, studied an aquatic single-celled organism called a cyanobacterium, which features a circadian clock with functions similar to a human’s. Their goal was to use the In Vitro Clock to examine what happens when the cyanobacterium’s clock resets at the molecular level, similar to how our circadian clocks undergo time zone changes during travel. Instead of collecting samples from in vitro reactions continuously for three to four days under the previous system, their new high-throughput method allowed them to immediately track results.
One of the most important real-time findings centered on the components of the circadian clock that are responsible for relaying the circadian rhythm from the core oscillator to gene expression. The researchers found that the elements that rhythmically modify a regulator to generate circadian gene expression—catalyzing enzymes called kinases—also play a crucial role in how the clock functions.
“For the first two decades after its discovery, most of the research has been centered on the core oscillator,” said Fang. “We now find that the kinases, previously thought to be just output components, are actually part of the whole clock.”
Fang said the discovery is similar to a concept rooted in physics called the observer effect in which the act of observation also influences the observed system. In this case, in order to gain the timing information (the act of observation) kept by the core oscillator, kinases bring disturbance to the core (the observed system).
“Kinases need to ask the core oscillator what time it is by physical interaction and thus they are affecting the core in perceptible ways,” said Golden, a professor and director of UC San Diego’s Center for Circadian Biology. “This is part of their natural function and now we see they’ve become part of the machine.”
The circadian clock controls the rhythmic expression of hundreds of genes in the cyanobacterium Synechococcus elongatus. The pacemaker of the clock is a core oscillator made up of three proteins: KaiA, KaiB and KaiC. Temporal information is relayed to a transcription factor, RpaA, via two kinases, CikA and SasA, that physically interact with the Kai complex.
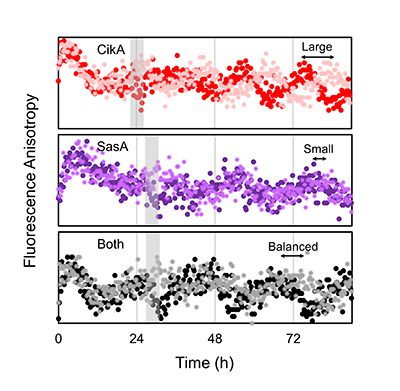 Real-time fluorescence measurements revealed that upon receiving the same resetting signal (indicated by gray bar), the In Vitro Clock system using only the CikA kinase shows a large response, while a SasA-only clock shows a small response. The clock system containing both kinases has a balanced response to the resetting signal, which is crucial for the cyanobacterium to properly align its internal clock with the environmental cycle.
Real-time fluorescence measurements revealed that upon receiving the same resetting signal (indicated by gray bar), the In Vitro Clock system using only the CikA kinase shows a large response, while a SasA-only clock shows a small response. The clock system containing both kinases has a balanced response to the resetting signal, which is crucial for the cyanobacterium to properly align its internal clock with the environmental cycle.
In fact, two kinases are required for a properly functioning circadian clock. Researchers who study circadian biology often refer to the core oscillator as the “gears” of the clock and the two kinases as the “hands” of the clock, both of which are needed to correctly tell time.
“We now know that the hands of the clock are actually part of the time-keeping mechanism,” said Golden. “If you don’t have both hands they don’t set time correctly because one of them is a stabilizer and one a perturber to the resetting signal, and you need both.”
Authors of the PNAS study include: Mingxu Fang, Archana Chavan, Andy LiWang and Susan Golden.
The research was supported by the National Institute of General Medical Sciences, National Institutes of Health (R35GM118290 and R35GM144110).
Publication: Mingxu Fang, et al., Synchronization of the circadian clock to the environment tracked in real time, PNAS (2023) DOI: 10.1073/pnas.2221453120
Original Story Source: UC San Diego

 Alerts Sign-up
Alerts Sign-up