Generative AI Imagines New Protein Structures
MIT researchers develop "FrameDiff," a computational tool that uses generative AI to craft new protein structures, with the aim of accelerating drug development and improving gene therapy.
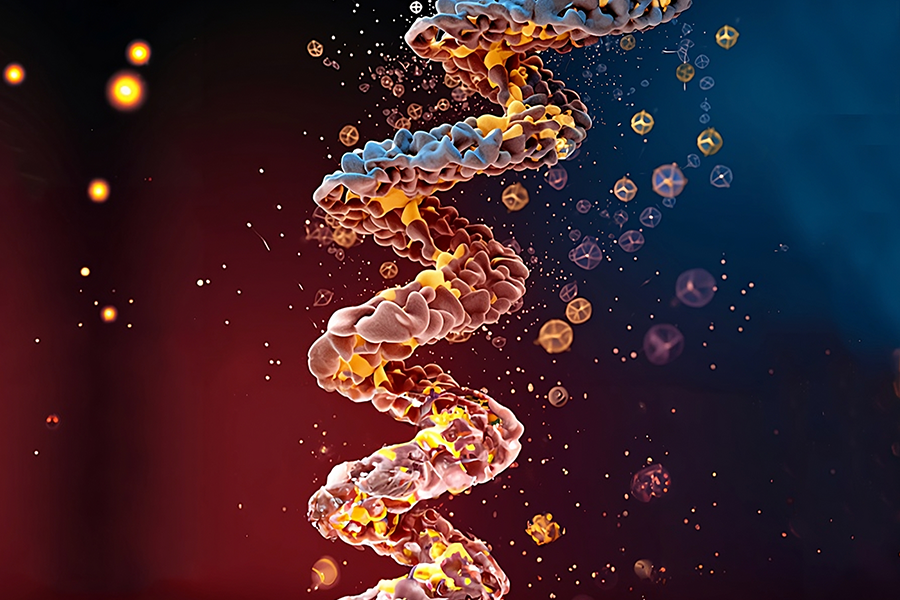 Biology is a wondrous yet delicate tapestry. At the heart is DNA, the master weaver that encodes proteins, responsible for orchestrating the many biological functions that sustain life within the human body. However, our body is akin to a finely tuned instrument, susceptible to losing its harmony. After all, we’re faced with an ever-changing and relentless natural world: pathogens, viruses, diseases, and cancer.
Biology is a wondrous yet delicate tapestry. At the heart is DNA, the master weaver that encodes proteins, responsible for orchestrating the many biological functions that sustain life within the human body. However, our body is akin to a finely tuned instrument, susceptible to losing its harmony. After all, we’re faced with an ever-changing and relentless natural world: pathogens, viruses, diseases, and cancer.
Imagine if we could expedite the process of creating vaccines or drugs for newly emerged pathogens. What if we had gene editing technology capable of automatically producing proteins to rectify DNA errors that cause cancer? The quest to identify proteins that can strongly bind to targets or speed up chemical reactions is vital for drug development, diagnostics, and numerous industrial applications, yet it is often a protracted and costly endeavor.
To advance our capabilities in protein engineering, MIT CSAIL researchers came up with “FrameDiff,” a computational tool for creating new protein structures beyond what nature has produced. The machine learning approach generates “frames” that align with the inherent properties of protein structures, enabling it to construct novel proteins independently of preexisting designs, facilitating unprecedented protein structures.
"In nature, protein design is a slow-burning process that takes millions of years. Our technique aims to provide an answer to tackling human-made problems that evolve much faster than nature's pace,” says MIT CSAIL PhD student Jason Yim, a lead author on a new paper about the work. “The aim, with respect to this new capacity of generating synthetic protein structures, opens up a myriad of enhanced capabilities, such as better binders. This means engineering proteins that can attach to other molecules more efficiently and selectively, with widespread implications related to targeted drug delivery and biotechnology, where it could result in the development of better biosensors. It could also have implications for the field of biomedicine and beyond, offering possibilities such as developing more efficient photosynthesis proteins, creating more effective antibodies, and engineering nanoparticles for gene therapy.”
Framing FrameDiff
Proteins have complex structures, made up of many atoms connected by chemical bonds. The most important atoms that determine the protein’s 3D shape are called the “backbone,” kind of like the spine of the protein. Every triplet of atoms along the backbone shares the same pattern of bonds and atom types. Researchers noticed this pattern can be exploited to build machine learning algorithms using ideas from differential geometry and probability. This is where the frames come in: Mathematically, these triplets can be modeled as rigid bodies called “frames” (common in physics) that have a position and rotation in 3D.
These frames equip each triplet with enough information to know about its spatial surroundings. The task is then for a machine learning algorithm to learn how to move each frame to construct a protein backbone. By learning to construct existing proteins, the algorithm hopefully will generalize and be able to create new proteins never seen before in nature.
Training a model to construct proteins via “diffusion” involves injecting noise that randomly moves all the frames and blurs what the original protein looked like. The algorithm’s job is to move and rotate each frame until it looks like the original protein. Though simple, the development of diffusion on frames requires techniques in stochastic calculus on Riemannian manifolds. On the theory side, the researchers developed “SE(3) diffusion” for learning probability distributions that nontrivially connects the translations and rotations components of each frame.
3D animation of a protein structure being generated from a bundle of purple dots
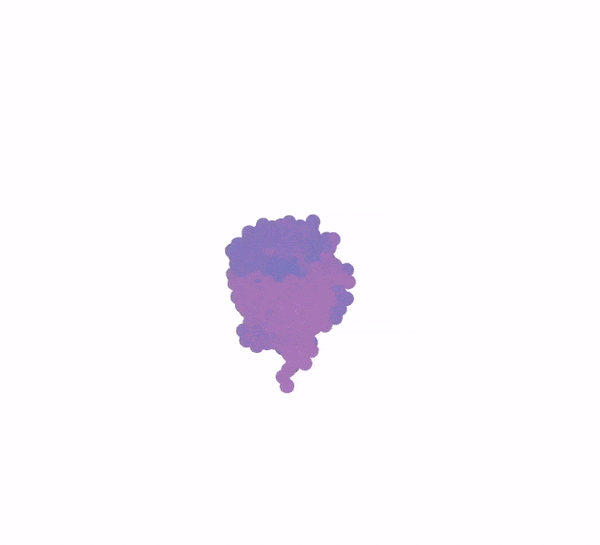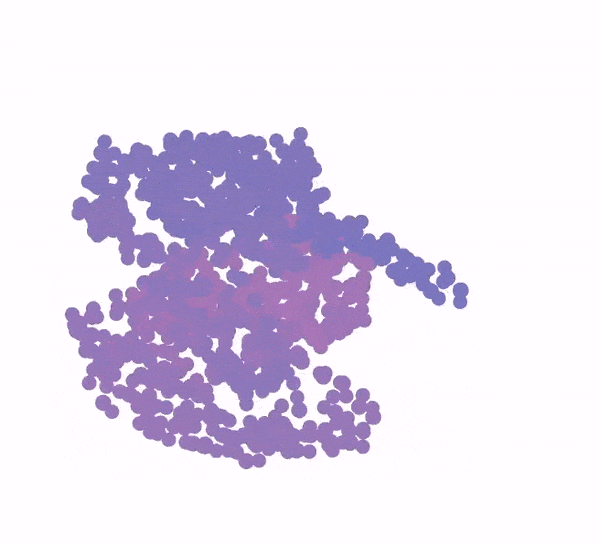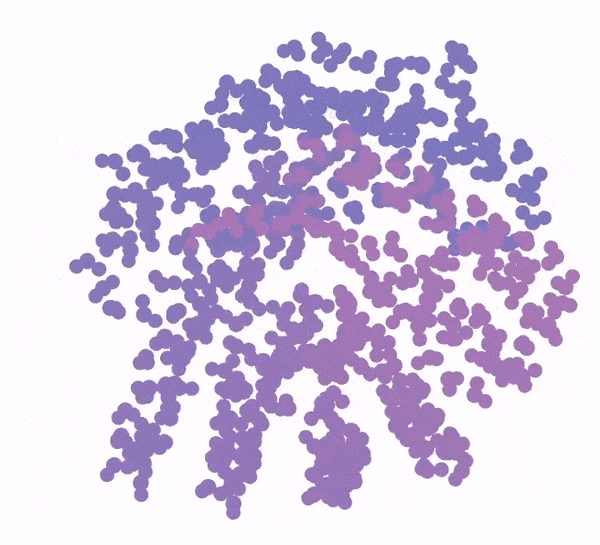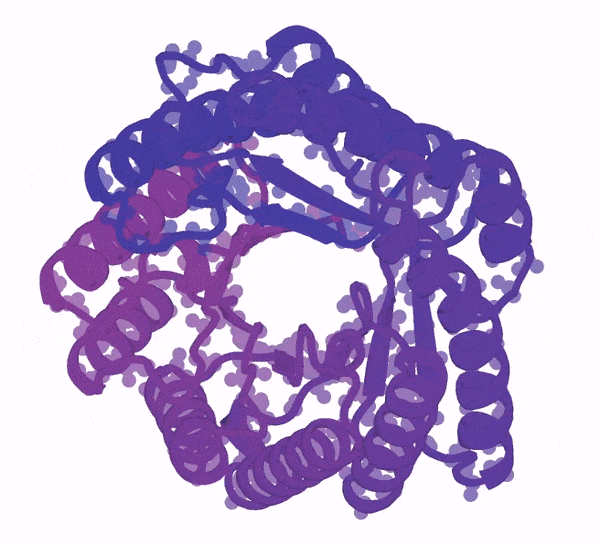 Generation of a protein structure with FrameDiff
Generation of a protein structure with FrameDiff
Image: Ian Haydon/Institute for Protein Design
The subtle art of diffusion
In 2021, DeepMind introduced AlphaFold2, a deep learning algorithm for predicting 3D protein structures from their sequences. When creating synthetic proteins, there are two essential steps: generation and prediction. Generation means the creation of new protein structures and sequences, while "prediction" means figuring out what the 3D structure of a sequence is. It’s no coincidence that AlphaFold2 also used frames to model proteins. SE(3) diffusion and FrameDiff were inspired to take the idea of frames further by incorporating frames into diffusion models, a generative AI technique that has become immensely popular in image generation, like Midjourney, for example.
The shared frames and principles between protein structure generation and prediction meant the best models from both ends were compatible. In collaboration with the Institute for Protein Design at the University of Washington, SE(3) diffusion is already being used to create and experimentally validate novel proteins. Specifically, they combined SE(3) diffusion with RosettaFold2, a protein structure prediction tool much like AlphaFold2, which led to “RFdiffusion.” This new tool brought protein designers closer to solving crucial problems in biotechnology, including the development of highly specific protein binders for accelerated vaccine design, engineering of symmetric proteins for gene delivery, and robust motif scaffolding for precise enzyme design.
Future endeavors for FrameDiff involve improving generality to problems that combine multiple requirements for biologics such as drugs. Another extension is to generalize the models to all biological modalities including DNA and small molecules. The team posits that by expanding FrameDiff's training on more substantial data and enhancing its optimization process, it could generate foundational structures boasting design capabilities on par with RFdiffusion, all while preserving the inherent simplicity of FrameDiff.
“Discarding a pretrained structure prediction model [in FrameDiff] opens up possibilities for rapidly generating structures extending to large lengths,” says Harvard University computational biologist Sergey Ovchinnikov. The researchers' innovative approach offers a promising step toward overcoming the limitations of current structure prediction models. Even though it's still preliminary work, it's an encouraging stride in the right direction. As such, the vision of protein design, playing a pivotal role in addressing humanity's most pressing challenges, seems increasingly within reach, thanks to the pioneering work of this MIT research team.”
Yim wrote the paper alongside Columbia University postdoc Brian Trippe, French National Center for Scientific Research in Paris' Center for Science of Data researcher Valentin De Bortoli, Cambridge University postdoc Emile Mathieu, and Oxford University professor of statistics and senior research scientist at DeepMind Arnaud Doucet. MIT professors Regina Barzilay and Tommi Jaakkola advised the research.
The team's work was supported, in part, by the MIT Abdul Latif Jameel Clinic for Machine Learning in Health, EPSRC grants and a Prosperity Partnership between Microsoft Research and Cambridge University, the National Science Foundation Graduate Research Fellowship Program, NSF Expeditions grant, Machine Learning for Pharmaceutical Discovery and Synthesis consortium, the DTRA Discovery of Medical Countermeasures Against New and Emerging threats program, the DARPA Accelerated Molecular Discovery program, and the Sanofi Computational Antibody Design grant. This research will be presented at the International Conference on Machine Learning in July.
Publication: Jason Yim, et al., SE(3) diffusion model with application to protein backbone generation, Cornell University (2023) DOI : 10.48550/arXiv.2302.02277
Original Story Source: Massachusetts Institute of Technology

 Alerts Sign-up
Alerts Sign-up