Chemical ‘Nose’ Sniffs Critical Differences In DNA Structures
Detection innovation could enhance drug development and cancer research
Small changes in the structure of DNA have been implicated in breast cancer and other diseases, but they’ve been extremely difficult to detect — until now.
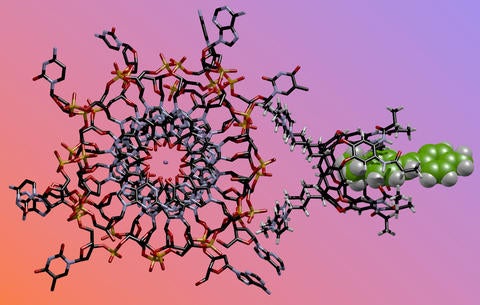 "Chemical nose" system used to detect G-quadruplex structures in DNA. (Richard Hooley/UCR)
"Chemical nose" system used to detect G-quadruplex structures in DNA. (Richard Hooley/UCR)
Using what they describe as a “chemical nose,” UC Riverside chemists are able to “smell” when bits of DNA are folded in unusual ways. Their work designing and demonstrating this system has been published in the journal Nature Chemistry.
“If a DNA sequence is folded, it could prevent the transcription of a gene linked to that particular piece of DNA,” said study author and UCR chemistry professor Wenwan Zhong. “In other words, this could have a positive effect by silencing a gene with the potential to cause cancer or promote tumors.”
Conversely, DNA folding could also have a negative effect.
“DNA folds could potentially keep viral proteins from being produced to minimize immune response,” Zhong said.
Studying how these folds might impact living beings, either positively or negatively, first requires scientists to detect their presence. To do that, UCR organic chemistry professor Richard Hooley and his colleagues modified a concept that has previously been used to sense other things, such as chemical components in different vintages of wine.
The chemicals in the system could be designed to look for nearly any kind of target molecule. However, the way the “nose” is typically used, it could not detect DNA. Only once Hooley’s group added additional, nonstandard components could the nose sniff out its DNA target.
“Humans detect smells by inhaling air containing odor molecules that bind to multiple receptors inside the nose,” Hooley explained. “Our system is comparable because we have multiple receptors able to interact with the DNA folds we’re looking for.”
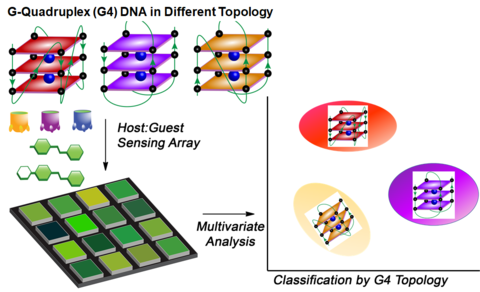 Illustration of the system responding to G-quadruplex detection. (Richard Hooley/UCR)
Illustration of the system responding to G-quadruplex detection. (Richard Hooley/UCR)
The chemical nose is composed of three parts: host molecules, fluorescent guest molecules, and DNA, which is the target. When the desired folds are present, the guest glows, alerting scientists to their presence in a sample.
DNA is made of four nucleic acids: guanine, adenine, cytosine and thymine. Most of the time, these acids form a double helix structure resembling a ladder. Guanine-rich regions sometimes fold in a different manner, creating what’s called a G-quadruplex.
Parts of the genome that form these quadruplex structures are extremely complex, though UC Riverside researchers have discovered that their folds are known to regulate gene expression, and they play a key role in keeping cells healthy.
For this experiment, the researchers wanted to demonstrate that they could detect one specific type of quadruplex composed of four guanines. Having done so, Zhong said the research team will try to build on their success.
“Now we think we can do more,” she said. “There are other three-dimensional structures in DNA, and we want to understand those as well.”
The researchers will examine how forces that damage DNA affect the ways they fold. They will also study RNA folding because RNA carries out important functions in a cell.
“RNA has even more complex structures than DNA, and is more difficult to analyze, but understanding its structure has great potential for disease research,” Zhong said
Publication: Junyi Chen, et al., Selective discrimination and classification of G-quadruplex structures with a host–guest sensing array, nature chemistry (2023). DOI: 10.1038/s41557-021-00647-9
Original Story Source: University of California – Riverside

 Alerts Sign-up
Alerts Sign-up