Modern Simulations Could Improve Mris
Rice engineers find more efficient models to analyze contrast agents that find disease
HOUSTON – (Sept. 20, 2021) – Gadolinium-based contrast agents, the gold standard in magnetic resonance imaging (MRI) to determine the health of a patient, can be improved, according to Rice University engineers who are refining models they first used to enhance oil and gas recovery.
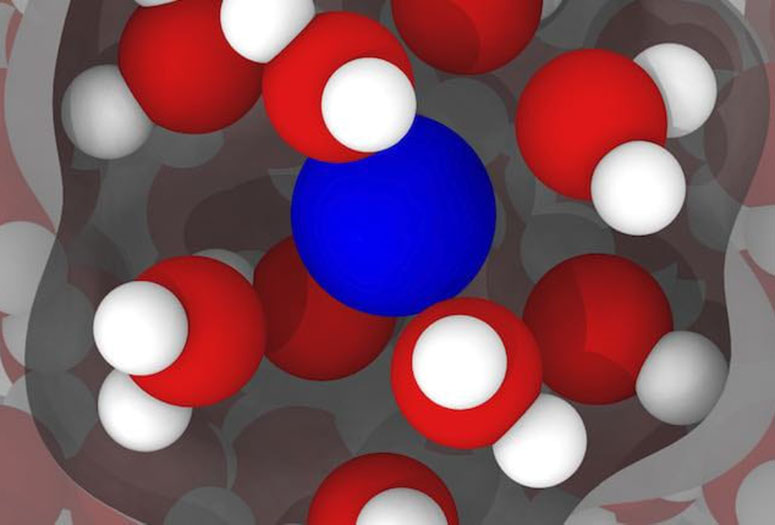 An illustration based on simulations by Rice engineers shows a gadolinium ion (blue) in water (red and white), with inner-sphere water -- the water most affected by the gadolinium -- highlighted. The researchers’ models of gadolinium in water show there’s room for improvement in compounds used as contrast agents in clinical magnetic resonance imaging. Illustration by Arjun Valiya Parambathu
An illustration based on simulations by Rice engineers shows a gadolinium ion (blue) in water (red and white), with inner-sphere water -- the water most affected by the gadolinium -- highlighted. The researchers’ models of gadolinium in water show there’s room for improvement in compounds used as contrast agents in clinical magnetic resonance imaging. Illustration by Arjun Valiya ParambathuThe team led by Dilip Asthagiri and Philip Singer of the George R. Brown School of Engineering had studied how nuclear magnetic resonance tools, commonly used in the oil industry to characterize deposits underground, could be optimized through molecular dynamics simulations.
“We addressed a lot of fundamental scientific questions there, and we wondered if there were other ways we could use these simulations,” Asthagiri said.
“There are roughly 100 million MRIs taken worldwide every year, and about 40% of them use gadolinium-based contrast agents, but the way they model MRI response to these agents hasn’t changed significantly since the 1980s,” Singer said. “We thought it would be a good test bed for our ideas.”
The results of their research appear in the Royal Society of Chemistry journal Physical Chemistry Chemical Physics.
Their paper demonstrates how limiting the number of parameters in simulations has the potential to improve the analysis of gadolinium-based contrast agents and how effective they are at imaging for clinical diagnosis. Their goal is to make better and more customizable contrast agents.
Doctors use MRI devices to “see” the state of soft tissues inside the body, including the brain, by inducing magnetic moments in the hydrogen nuclei of ever-present water molecules to align along the magnetic field. The device detects bright spots when the aligned nuclei “relax” back to thermal equilibrium following an excitation, and the faster they relax, the brighter the contrast.
That’s where paramagnetic gadolinium-based contrast agents come in. “Gadolinium ions increase sensitivity and make the signal brighter by decreasing the T1 relaxation time of hydrogen nuclei,” Asthagiri said. “Our ultimate goal is to help the optimization and design of these agents.”
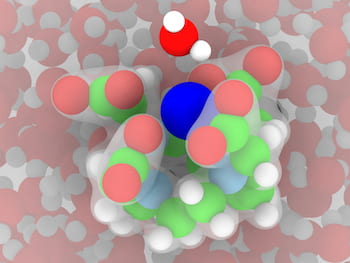 An illustration based on simulations by Rice University engineers shows a gadolinium ion (dark blue) surrounded by a chelate known as DOTA in water. The chelate is required to minimize gadolinium retention in the body after a magnetic resonance imaging scan. The green atoms are carbon and light blue are nitrogen. Illustration by Arjun Valiya Parambathu
An illustration based on simulations by Rice University engineers shows a gadolinium ion (dark blue) surrounded by a chelate known as DOTA in water. The chelate is required to minimize gadolinium retention in the body after a magnetic resonance imaging scan. The green atoms are carbon and light blue are nitrogen. Illustration by Arjun Valiya ParambathuTypically, gadolinium is “chelated” -- surrounded by metal ions -- to make it less toxic. “The body doesn’t remove gadolinium by itself and needs to be chelated so the kidneys can get rid of it after a scan,” Singer said. “But chelation also slows the molecular rotation, and that creates better contrast in the MRI image.”
The researchers noted “chelate” comes from the Greek word for claw. “In this case, these claws grip the gadolinium to make it stable,” he said. “We hope our models help us design a stronger grip, which will make them safer while maximizing their ability to increase contrast.”
They acknowledged that gadolinium chelates, which revolutionized MRI testing when introduced in the late 1980s, have been controversial lately since it was discovered that patients with kidney impairment were unable to eliminate all of the toxins. “They’ve since worked out that if you have good kidney function, the benefits outweigh the potential risks,” Singer said.
The team is also adapting its models beyond interactions with water. “In biological systems, cells have other constituents like osmolytes and denaturants like urea, so we’re modeling gadolinium with these different environments to build toward a variety of applications,” Asthagiri said.
 Philip Singer
Philip Singer Dilip Asthagiri
Dilip AsthagiriCo-authors of the paper are Rice graduate students Arjun Valiya Parambathu, Thiago Pinheiro dos Santos and Yunke Liu; senior research scientist Lawrence Alemany; George Hirasaki, the A.J. Hartsook Professor Emeritus and a research professor; and Walter Chapman, the William W. Akers Professor of Chemical and Biomolecular Engineering.
Vinegar Technologies LLC, Chevron Energy Technology, the Rice University Consortium on Processes in Porous Media, the Department of Energy Office of Science and the Texas Advanced Computing Center at the University of Texas at Austin supported the research
Publication: Philip M. Singer, et al., Predicting 1H NMR relaxation in Gd3+-aqua using molecular dynamics simulations, Physical Chemistry Chemical Physics (2021). DOI: 10.1039/D1CP03356E
Original Story Source: Rice University

 Alerts Sign-up
Alerts Sign-up